Jostine Joby presented his research in a faculty seminar!
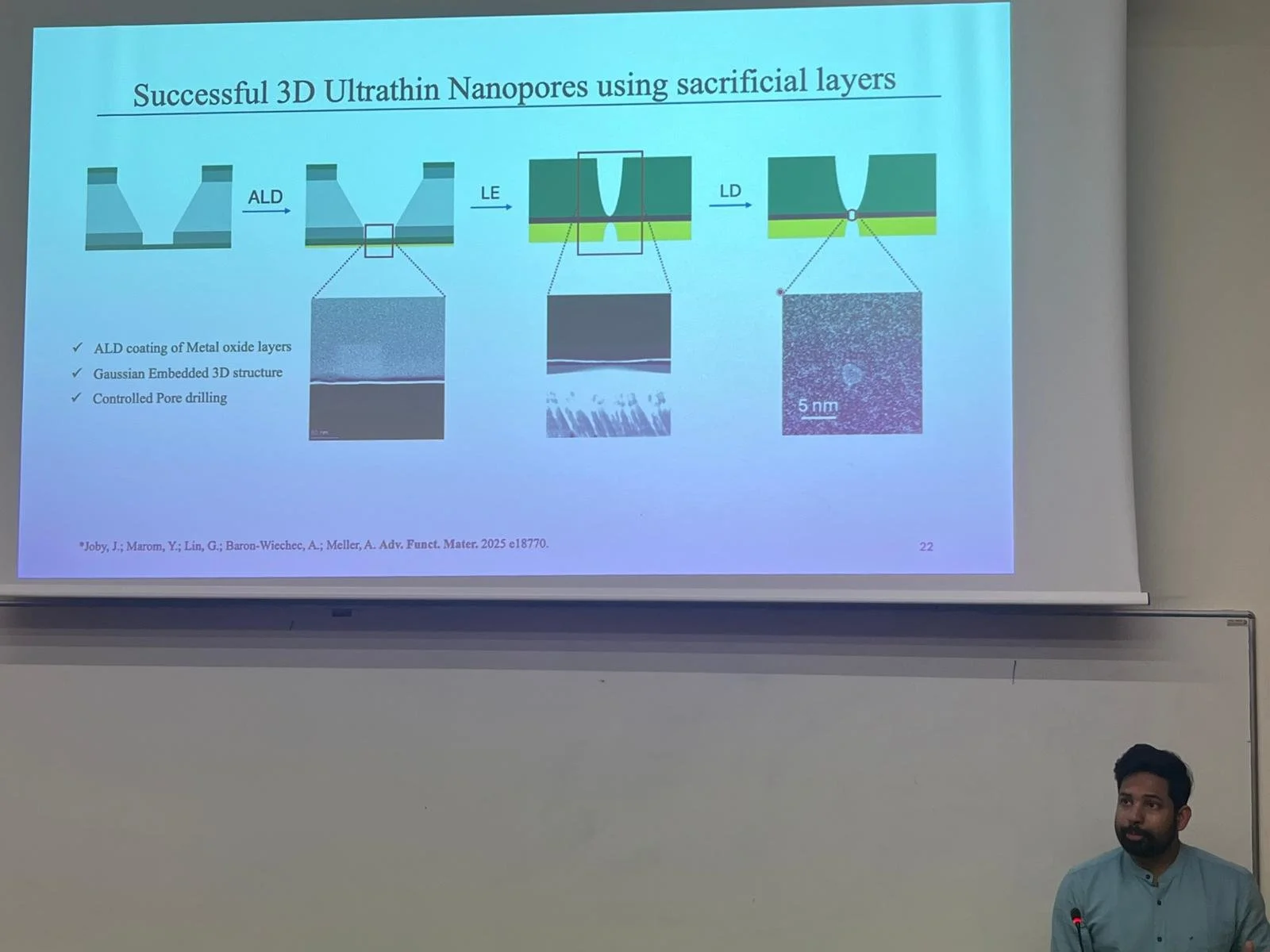
In his seminar, Jostine Joby, Lab PhD student, discussed his innovative work on ultrathin metal-oxide nanopores for single-molecule detection. Using a combination of laser sculpturing in liquid and atomic layer deposition, Jostine’s approach allows rapid, label-free detection of DNA, microRNA, and proteins with exceptional precision.
This technique provides a detailed analysis of individual biomolecules, offering a fast, cost-effective, and highly accurate tool with strong potential for next-generation biosensing and single-molecule sequencing.
Great work, Jostine!
New Paper by Jostin Joby Published in Advanced Functional Materials
Congratulations to our PhD student Jostin Joby on his recent publication in Advanced Functional Materials!
The study presents a novel laser-based method for creating ultrathin metal-oxide nanopores that can detect and distinguish individual biomolecules with exceptional precision. By combining laser sculpturing in liquid with atomic layer deposition, the team achieved rapid, label-free detection of DNA, microRNA, and proteins — paving the way for next-generation biosensing and single-molecule sequencing technologies.
Lab Fun Day & Farewell
Yesterday, our lab team spent a wonderful day together in Haifa!
We joined a guided food tour in the lively Talpiot Market, tasted delicious dishes, and enjoyed learning about the local food culture. It was a great chance to relax, laugh, and spend time together outside the lab.
Later, we also said goodbye to Barak, Noam, and Malak, who are leaving the lab to start new adventures. We will really miss them and wish them all the best!
The day was full of smiles, good food, and great company. A perfect way to celebrate our amazing team!
Our Lab’s New Paper, Led by Postdoctoral Fellow Neeraj Soni, Unveiling a Novel Strategy for Single-Molecule Protein Analysis
In this work, we demonstrate that small oligonucleotides can act as proxies for cysteine residues to ultimately reveal protein identity!
We further show how protein translocation speed can be regulated to enhance time resolution and how the delivery of single-file proteins is significantly improved using negatively charged oligonucleotides. Finally, we apply a machine learning approach to identify specific proteins within a mixture, highlighting the potential of this strategy for advancing protein fingerprinting and single-molecule analysis.
Click here to read the article
Prof. Meller preseting the lab resent work at the CECAM workshop in Cagliari, italy
Prof. Amit Meller presented our lab’s recent work at the CECAM workshop, “Nanopores: From Basics to Applications.”
His talk focused on the use of solid-state nanopores for label-free identification of mitochondrial and cell-free DNA, as well as for protein fingerprinting.
Read more on CECAM website
Meller Lab Beach Gathering
Our team enjoyed a fun and relaxing day at the beach, filled with sunshine, good food, and great company. It was a perfect break from the lab and a chance to connect outside work. Thanks to everyone who joined!