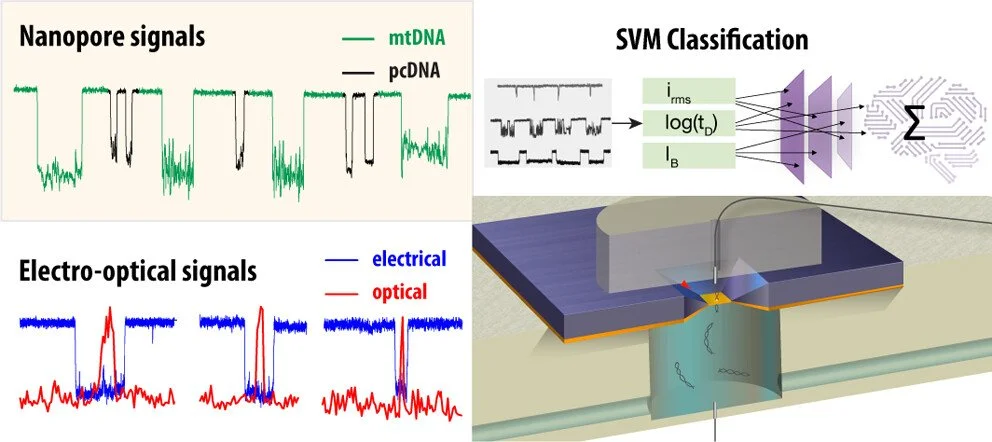
Machine Learning Enables Rapid Detection of Unamplified mtDNA
We are excited to share groundbreaking research from out lab. Recently featured on the university's website, showcases an innovative approach for detecting mitochondrial DNA (mtDNA) without the need for amplification—a significant advancement in the fields of diagnostics and genomics.
Our innovative nanotechnology enables direct analysis of native mitochondrial DNA without amplification, dramatically improving both speed and accuracy. This breakthrough will accelerate research into mitochondrial dysfunctions linked to cancer and neurodegenerative diseases.
Malak Hijazi presented her research in faculty Seminar title : “ Studying fragmentation distribution of cell free DNA(cfDNA) in healthy and sick patients using solid-state nanopores”
In her seminar, Malak discussed the use of solid-state nanopores to detect cell-free DNA (cfDNA) length variations, a promising approach for non-invasive disease monitoring through liquid biopsy.
Solid-state nanopores enable direct, label-free, single-molecule analysis of cfDNA fragment sizes without the need for amplification, avoiding biases common in qPCR and NGS. By analyzing ion-current signatures during translocation, this method reveals detailed size distributions, enhancing diagnostic accuracy. It offers a fast, cost-effective tool with strong potential for early detection and point-of-care use in oncology.
Great work Malak!
Check out our review in Analytical Chemistry on The Emergence of Nanofluidics for Single-Biomolecule Manipulation and Sensing
In this paper, we explore how tiny fluid channels—called nanochannels—are being used to study individual molecules like DNA and proteins. These nanofluidic devices can stretch and organize molecules, making their detection more precise. They are already used for genome mapping and showing great promise for identifying and studying single proteins, which will revolutionize next-generation medical tests.
Read the full paper here: https://pubs.acs.org/doi/10.1021/acs.analchem.4c06684
Check our new paper in ACS Nano Journal about amplification-free mitochondrial DNA quantification
In our latest research, we introduce a solid-state nanopore method for label-free, amplification-free detection of mitochondrial DNA (mtDNA), a key biomarker for disease. Using machine learning and exonuclease digestion, we achieve high accuracy in distinguishing mtDNA from genomic DNA in biological samples. This approach offers picomolar sensitivity with minimal preparation, making it ideal for clinical diagnostics.
Noam presented his final PhD seminar entitled "Single-Molecule Protein Sensing for Early AMD Diagnosis”
In his seminar, he talked about the use of single-molecule sensing technology for early detection of age-related macular degeneration (AMD), a leading cause of vision loss in older adults. Unlike traditional methods that detect physical damage, this approach analyzes molecular biomarkers like VEGF and Clusterin with high precision. Single-molecule techniques, such as nanopore-based sensors or dynamic sensing using nanoparticles, allow real-time tracking of individual protein interactions, enabling ultrasensitive detection at subfemtomolar levels. These methods improve diagnostic accuracy by identifying molecular changes before significant vision loss occurs, paving the way for earlier intervention and personalized treatments. This innovation could transform AMD management, reducing its socioeconomic impact and enhancing patient outcomes.
Welcoming Jiban Mondal to our Lab!
Jiban Mondal, new PhD student joined our Lab.
Jiban’s research will focus on developing a plasmonic nanopore system to enhance optical signal detection for full-length protein sensing. His work will integrate opto-electrical data collection (OptiPore) to improve signal resolution and enable more precise protein characterization.
Welcome to our team, Jiban, and best of luck!