2024
Full-length single protein molecules tracking and counting inthin silicon channels
Shilo Ohayon, Liran Taib, Navneet C. Verma, Marzia Iarossi, Ivy Bhattacharya, Barak Marom, Diana Huttner, Amit Meller, Advanced Materials. 2024, DOI: https://doi.org/10.1002/adma.202314319
2023
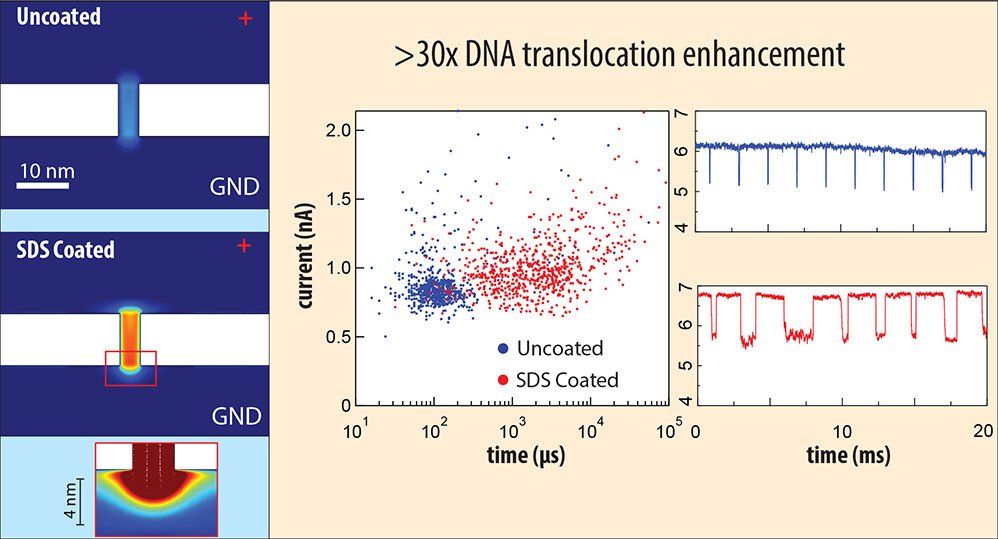
Over 30-Fold Enhancement in DNA Translocation Dynamics through Nanoscale Pores Coated with an Anionic Surfactant
Neeraj Soni, Navneet Chandra Verma, Noam Talor, and Amit Meller, Nano Lett. 2023, DOI: https://doi.org/10.1021/acs.nanolett.3c01096
2022
Nanopore-based technologies beyond DNA sequencing
Yi-Lun Ying, Zheng-Li Hu, Shengli Zhang, Yujia Qing, Alessio Fragasso, Giovanni Maglia, Amit Meller, Hagan Bayley, Cees Dekker and Yi-Tao Long,
Nature Nanotechnology, 2022, DOI: https://doi.org/10.1038/s41565-022-01193-2
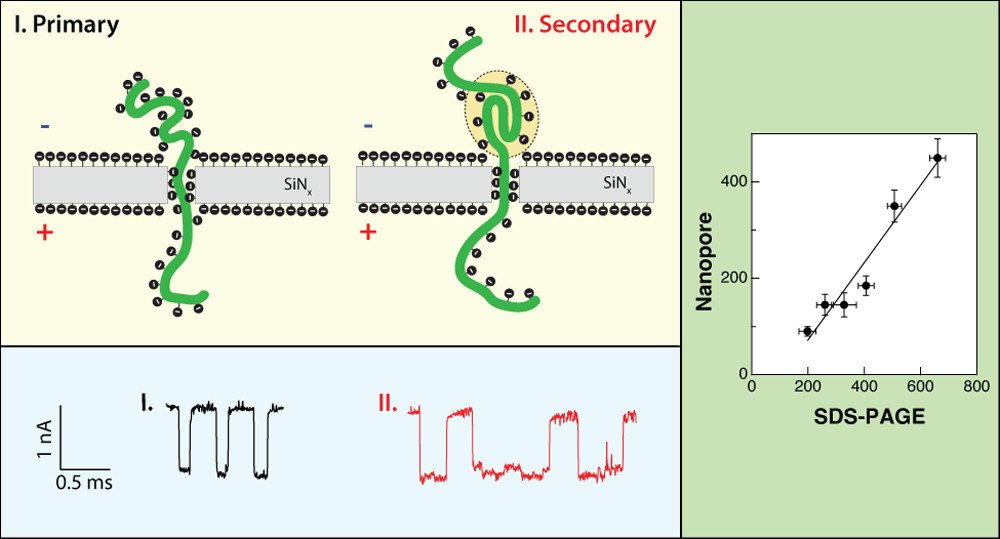
Single-File Translocation Dynamics of SDS-Denatured, Whole Proteins through Sub-5 nm Solid-State Nanopores
Neeraj Soni, Noam Freundlich, Shilo Ohayon, Diana Huttner, and Amit Meller,
ACS Nano, 2022, DOI: 10.1021/acsnano.2c05391
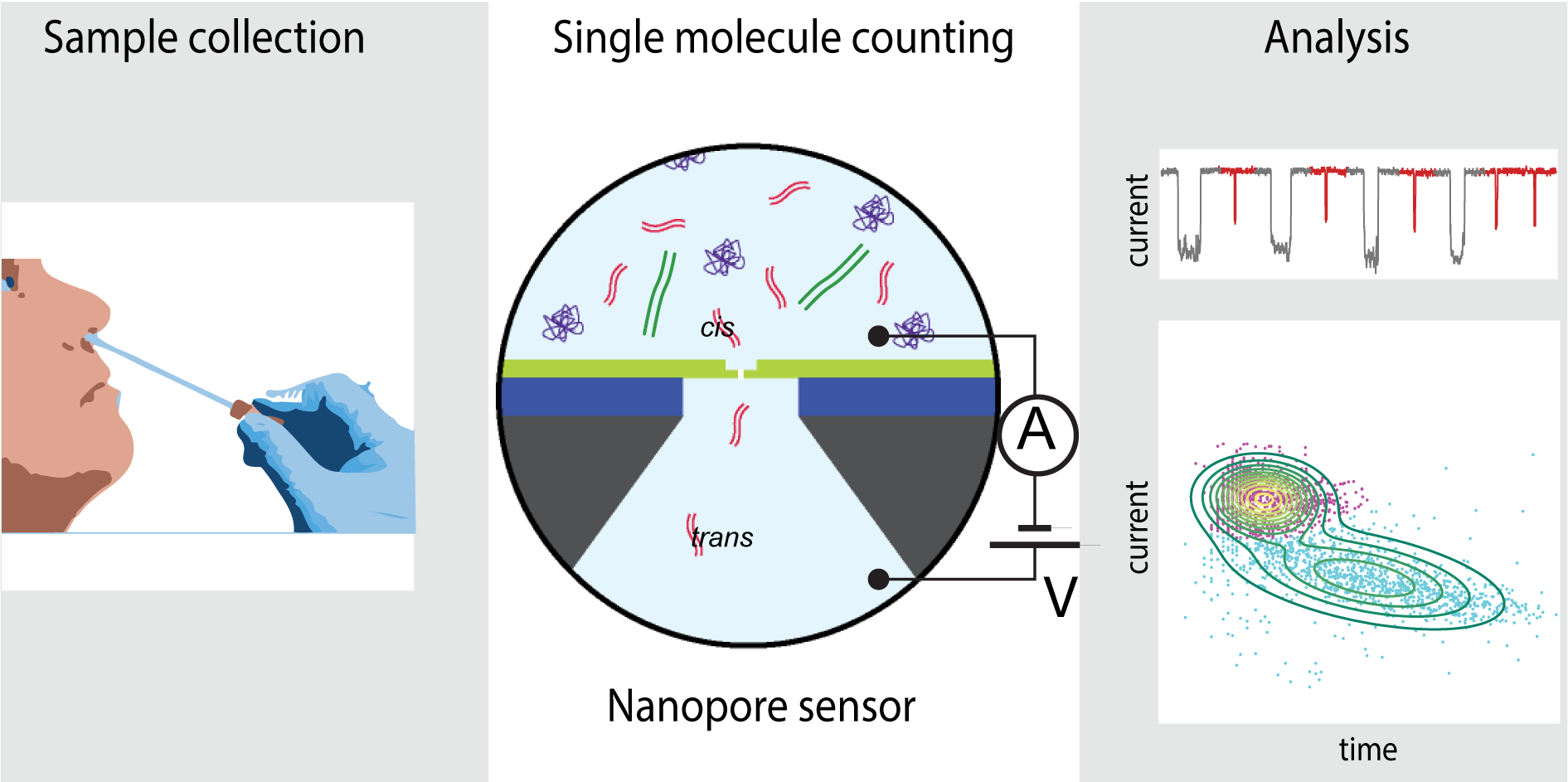
Purely electrical SARS-CoV-2 sensing based on single-molecule counting
Xander F. van Kooten, Yana Rozevsky, Yulia Marom, Efrat Ben Sadeh and Amit Meller, Nanoscale, 2022, DOI: 10.1039/D1NR07787B.
Are nanopore technologies ready for the proteomic challenge primetime?
Amit Meller, Molecular Cell 2022, Volume 82, Issue 2, 2021.12.018
2021
Lifetime-based analysis of binary fluorophores mixtures in the low photon count limit
Maisa Nasser, Amit Meller, iScience, 2021, ISCI 103554.
Fast and Deterministic Fabrication of Sub-5 Nanometer Solid-State Pores by Feedback-Controlled Laser Processing
Eran Zvuloni, Adam Zrehen, Tal Gilboa, Amit Meller, ACS Nano, 2021, acsnano.1c03773.
The emerging landscape of single-molecule protein sequencing technologies
Javier A. Alfaro et. al. Nature Methods. 2021. 10.1038.
Nanopore identification of single nucleotide mutations in circulating tumor DNA by multiplexed ligation
Nitza Burck, Tal Gilboa, Abhilash Gadi, Michelle Patkin Nehrer, Robert J. Schneider and Amit Meller, Clin. Chem. 2021.
2020
On‑chip protein separation with single‑molecule resolution
A. Zrehen, S. Ohayon, D. Huttner and A. Meller, Sci Rep 2020, 10-15313–12.
Quantification of mRNA Expression Using Single-Molecule Nanopore Sensing
Y. Rozevsky, T. Gilboa, X. F. van Kooten, D. Kobelt, D. Huttner, U. Stein and A. Meller, ACS Nano, 2020, acsnano.0c06375.
Microfluidic device for coupling isotachophoretic sample focusing with nanopore single-molecule sensing
J. Spitzberg, X. F. van Kooten, M. Bercovici and A. Meller, Nanoscale, 2020, DOI:10.1039/D0NR05000H.
Sub-second, super-resolved imaging of biological systems using parallel EO-STED
Girsault A. and Meller A. Optics Lett. 2020; 10.1364/OL.392822
2019
On-Chip Stretching, Sorting, and Electro-Optical Nanopore Sensing of Ultralong Human Genomic DNA
Zrehen, A., Huttner, D., Meller, A. ACS Nano. 2019 : acsnano.9b07873.
Scanning Nanopore Microscopy - A new tool for cell signaling research
Meller A. Nature Nanotechnology 2019,
DOI: 10.1038/s41565-019-0505-z
Simulation of single-protein nanopore sensing shows feasibility for whole-proteome identification
Ohayon, S., Girsault, A., Nasser, M., Shen-Orr, S. & Meller, A. Simulation of single-protein nanopore sensing shows feasibility for whole-proteome identification. PLoS Comput. Biol. 2019, 15, e1007067–21.
Automated, Ultra-Fast Laser-Drilling of Nanometer Scale Pores and Nanopore Arrays in Aqueous Solutions
Gilboa, T.; Zvuloni, E.; Zrehen, A.; Squires, A.H.; and Meller A. Automated, Ultra-Fast Laser-Drilling of Nanometer Scale Pores and Nanopore Arrays in Aqueous Solutions. Adv. Funct. Mater. 2019, 1900642.
Plasmonic-Nanopore Biosensors for Superior Single-Molecule Detection
Spitzberg, J.D.; Zrehen, A.; van Kooten, X.F. and Meller, A. Plasmonic‐Nanopore Biosensors for Superior Single‐Molecule Detection. Adv Mater. 2019, 313: 1900422–18. doi:10.1002/adma.201900422
2018
A Solid‐State Hard Microfluidic–Nanopore Biosensor with Multilayer Fluidics and on‐Chip Bioassay/Purification Chamber.
Varongchayakul, N.; Hersey, J. S.; Squires, A. H.; Meller, A.; Grinstaff, M. W. A Solid‐State Hard Microfluidic–Nanopore Biosensor with Multilayer Fluidics and on‐Chip Bioassay/Purification Chamber. Adv. Funct. Mater. 2018, 1804182.
Single-Molecule Discrimination of Labeled DNAs and Polypeptides Using Photoluminescent-Free TiO2 Nanopores
Wang, R.; Gilboa, T.; Song, J.; Huttner, D.; Grinstaff, M.W.; Meller, A. Single-Molecule Discrimination of Labeled DNAs and Polypeptides Using Photoluminescent-Free TiO2 Nanopores. ACS Nano, 2018, DOI: 10.1021/acsnano.8b07055
Optically-Monitored Nanopore Fabrication Using a Focused Laser Beam
Gilboa, T.; Zrehen, A.; Girsault, A.; Meller, A. Optically-Monitored Nanopore Fabrication Using a Focused Laser Beam. Sci Rep 2018, 8, 9765.
Single-molecule protein sensing in a nanopore: a tutorial
Varongchayakul, N.; Song, J.; Meller, A.; Grinstaff, M. W. Single-molecule protein sensing in a nanopore: a tutorial. Chem Soc Rev, 2018
Sensing Native Protein Solution Structures Using a Solid-State Nanopore: Unraveling the States of VEGF
Varongchayakul, N.; Huttner, D.; Grinstaff, M. W.; Meller, A.. Sensing Native Protein Solution Structures Using a Solid-State Nanopore: Unraveling the States of VEGF. Sci Rep, 2018, 8:1017, 1-9.
2017
Single-Molecule DNA Unzipping Reveals Asymmetric Modulation of a Transcription Factor by Its Binding Site Sequence and Context.
Rudnizky, S., Khamis, H., Malik, O., Squires, A. H.; Meller A ., Melamed,. Kaplan, A. (2017) Single-Molecule DNA Unzipping Reveals Asymmetric Modulation of a Transcription Factor by Its Binding Site Sequence and Context. Nuc. Acid Res.
Real-time visualization and sub-diffraction limit localization of nanometer-scale pore formation by dielectric breakdown
Zrehen, A; Gilboa, T. and Meller, A. (2017) Real-time visualization and sub-diffraction limit localization of nanometer-scale pore formation by dielectric breakdown, Nanoscale, 9(42), 16437-16445. DOI: 10.1039/c7nr02629c
Light-Enhancing Plasmonic-Nanopore Biosensor for Superior Single-Molecule Detection.
Assad, O. N.; Gilboa, T.; Spitzberg, J.; Juhasz, M.; Weinhold, E.; Meller, A. (2017) Light-Enhancing Plasmonic-Nanopore Biosensor for Superior Single-Molecule Detection. Adv. Mater., 1605442–1605449.
Single-Molecule Characterization of DNA-Protein Interactions Using Nanopore Biosensors
Review - Squires, A. H.; Gilboa, T.; Torfstein, C.; Varongchayakul, N.; Meller, A. Single-Molecule Characterization of DNA-Protein Interactions Using Nanopore Biosensors; Methods in Enzymol. Part B, 1-33.; 2017, Elsevier Inc., ; pp. 1–33.
2015-2016
Single-Molecule DNA Methylation Quantification Using Electro-Optical Sensing in Solid-State Nanopores.
Gilboa, T.; Torfstein, C.; Juhasz, M.; Grunwald, A.; Ebenstein, Y.; Weinhold, E.; Meller, A. Single-Molecule DNA Methylation Quantification Using Electro-Optical Sensing in Solid-State Nanopores. ACS Nano, 2016, 10, 8861–8870. doi:10.1021/acsnano.6b04748
Functionalized Nanofiber Meshes Enhance Immunosorbent Assays.
Hersey, J. S., Meller, A. & Grinstaff, M. W. Functionalized Nanofiber Meshes Enhance Immunosorbent Assays. Anal. Chem. acs.analchem.5b03386 (2015). doi:10.1021/acs.analchem.5b03386
Genomic Pathogen Typing Using Solid-State Nanopores
Squires, A., Atas, E. and Meller, A. Genomic Pathogen Typing Using Solid-State Nanopores, PLoS ONE. 2015, 10, e0142944.
Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores
Nir, I., Huttner, D. and Meller, A. Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores, Biophys. J. 108(9), 2015, 2340-2349.
Nanopore sensing of individual transcription factors bound to DNA
Squires, A., Atas, E. and Meller, A. Nanopore sensing of individual transcription factors bound to DNA, Sci. Rep. 2015, 5, 1-11.
Two Color DNA Barcode Detection in Photoluminescence Supressed Silicon Nitride Nanopores
Assad, O.N., Di Fiori, N., Squires, A.H. and Meller, A. Two Color DNA Barcode Detection in Photoluminescence Supressed Silicon Nitride Nanopores, Nano Letters, 2015, 15, 745-752. DOI: 10.1021/nl504459c
Optical sensing and analyte manipulation in solid- state nanopores
Review - Hitron, T. and Meller A. Optical sensing and analyte manipulation in solid- state nanopores. Analyst. 2015, DOI: 10.1039/c4an02388a
2014
Probing Solid-State Nanopores with Light for the Detection of Unlabeled Analyte
Anderson, B.N., Assad, O.N., Gilboa,T., Squires, A.H. Bar, D. and Meller, A. Probing Solid-State Nanopores with Light for the Detection of Unlabeled Analyte, ACS Nano, 2014, 8(11), 11836-11845.
Stationary nanoliter droplet array with a substrate of choice for single adherent/nonadherent cell incubation and analysis
Shemesh, J., Ben Arye, T., Avesar,J., Kang,J.H., Fine,A., SuperM., Meller, A., Ingber, D.E. and Levenberg S. Stationary nanoliter droplet array with a substrate of choice for single adherent/nonadherent cell incubation and analysis, Proc. Natl. Acad. Sci. USA, 2014, 111(31), 11293-11298.
Single Moleculae Kinetics of the Eukaryotic Initiation Factor 4AI Upon RNA Unwinding
Sun, Y., Atas, E., Lindqvist, L .M., Sonenberg, N., Pelletier, J. and A. Meller, Single Moleculae Kinetics of the Eukaryotic Initiation Factor 4AI Upon RNA Unwinding, Structure, 2014, 22(7), 941-948.
2011-2013
Optoelectronic control of surface charge and translocation dynamics in solid-state nanopores
Di Fiori, N. Squires, A. H, Bar, D., Gilboa, T. Moustakas, T.D. and A. Meller. Optoelectronic control of surface charge and translocation dynamics in solid-state nanopores, Nature Nanotechnology, 2013, 8, 946-951.
A Nanopore–Nanofiber Mesh Biosensor To Control DNA Translocation
Squires, A. H., J. S. Hersey, M. W. Grinstaff, and A. Meller. A Nanopore–Nanofiber Mesh Biosensor To Control DNA Translocation . J. Am. Chem. Soc., 2013, 135(44), 16304-116307
pH Tuning of DNA Translocation Time through Organically Functionalized Nanopores
Anderson BN, Muthukumar M, Meller A. pH Tuning of DNA Translocation Time through Organically Functionalized Nanopores. ACS Nano. 2012, 7(2): 1408-1414
Fabrication and characterization of solid-state nanopore arrays for high-throughput DNA sequencing
Dela Torre R, Larkin J, Singer A, Meller A. Fabrication and characterization of solid-state nanopore arrays for high-throughput DNA sequencing. Nanotechnology. 2012, 23(38):385308. doi: 10.1088/0957-4484/23/38/385308. Epub 2012 Sep 5.
The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase
Sun Y, Atas E, Lindqvist L, Sonenberg N, Pelletier J, Meller A. The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucleic Acids Res. 2012, 40(13), 6199-207. doi: 10.1093/nar/gks278. Epub 2012 Mar 28.
Nanopore detachment kinetics of poly(A) binding proteins from RNA molecules reveals the critical role of C-terminus interactions
Lin J, Fabian M, Sonenberg N, Meller A. Nanopore detachment kinetics of poly(A) binding proteins from RNA molecules reveals the critical role of C-terminus interactions. Biophys J. 2012, 102(6), 1427-1434. doi: 10.1016/j.bpj.2012.02.025. Epub 2012 Mar 20.
Electronic barcoding of a viral gene at the single-molecule level
Singer A, Rapireddy S, Ly DH, Meller A. Electronic barcoding of a viral gene at the single-molecule level. Nano Lett. 2012, 12(3):1722-1728. doi: 10.1021/nl300372a. Epub 2012 Feb 24.
Programmed trapping of individual bacteria using micrometre-size sieves
Kim, M.-C., Isenberg, B.C., Sutin, J., Meller, A., Wong, J.Y., and Klapperich, C.M. Programmed trapping of individual bacteria using micrometre-size sieves. Lab on a Chip 2011, 11, 1089-1095.
DNA Capture and Translocation through Nanoscale Pores - a Fine Balance of Electrophoresis and Electroosmosis
Review - Squires A.H. and Meller A. DNA Capture and Translocation through Nanoscale Pores - a Fine Balance of Electrophoresis and Electroosmosis, Biophysical J. New and Notable, 2013, 105, 543-544.
DNA sequencing by nanopore-induced photon emission
Review - Singer A, McNally B, Torre RD, Meller A. DNA sequencing by nanopore-induced photon emission. Methods Mol Biol. 2012, 870:99-114. doi: 10.1007/978-1-61779-773-6_6.
DNA sequencing and bar-coding using solid-state nanopores
Review - Atas E, Singer A, Meller A. DNA sequencing and bar-coding using solid-state nanopores. Electrophoresis. 2012, 33(23):3437-47. doi: 10.1002/elps.201200266. Epub 2012 Oct 30.
2010
Mechanisms governing the control of mRNA translation
Livingstone, M., Atas, E., Meller, A., and Sonenberg, N. Mechanisms governing the control of mRNA translation. Phys Biol, 2010, 7, 021001.
Detection of urea-induced internal denaturation of dsDNA using solid-state nanopores
Singer, A., H. Kuhn, M. Frank-Kamenetskii, and A. Meller. Detection of urea-induced internal denaturation of dsDNA using solid-state nanopores J. Phys. Cond-Mat., 2010, 22, 454111.
Optical Recognition of Converted DNA Nucleotides for Single-Molecule DNA Sequencing Using Nanopore Arrays
McNally, B., A. Singer, Z. Yu, Y. Sun, Z. Weng, and A. Meller. Optical Recognition of Converted DNA Nucleotides for Single-Molecule DNA Sequencing Using Nanopore Arrays. Nano Letters, 2010, 10(6) 2237-44.
The effect of dye-dye interactions on the spatial resolution of single-molecule FRET measurements in nucleic acids
Di Fiory, N. and A. Meller. The effect of dye-dye interactions on the spatial resolution of single-molecule FRET measurements in nucleic acids. Biophys. J., 2010, 98, 2265-72
Helix-coil kinetics of individual polyadenylic acid molecules in a protein channel
Lin, J., Kolomeisky, A. and Meller, A. Helix-coil kinetics of individual polyadenylic acid molecules in a protein channel. Phys. Rev. Lett. 2010, 104, 158101-4
Nanopore-based sequence-specific detection of duplex DNA for genomic profiling
Singer, A., Wanunu, M., Morrison, W., Kuhn, H., Frank-Kamenetskii, M., and Meller, A. Nanopore-based sequence-specific detection of duplex DNA for genomic profiling. Nano Letters. 2010, 10, 738-742.
Synchronous optical and electrical detection of bio-molecules traversing through solid-state nanopores
Soni, V. G., Singer, A., Yu, Z., Sun, Y., McNally, B. and Meller, A. Synchronous optical and electrical detection of bio-molecules traversing through solid-state nanopores. Rev. Sci. Instru. 2010, 81, 014301-014307.
Electrostatic Focusing of Unlabeled DNA into Nanoscale Pores using a Salt Gradient
Wanunu, M., Morrison, W., Rabin, Y., Grosberg, A. Y. and Meller, A. Electrostatic Focusing of Unlabeled DNA into Nanoscale Pores using a Salt Gradient, Nature Nano. 2010, 5,160-165.
Localized Joule heating produced by ion current focusing through micron-size holes.
Viasnoff, V., U. Bockelmann, A. Meller, H. Isambert, L. Laufer, and Y. Tsori. Localized Joule heating produced by ion current focusing through micron-size holes. Appl. Phys. Lett. 2010, 96, 163701-163703.
Nanopore-based Sensing of Individual Nucleic Acid Complexes
Review - Singer, A. and A. Meller. (2010). Nanopore-based Sensing of Individual Nucleic Acid Complexes. Israel Journal of Chemistry 49:323–331.
Nanopore Force Spectroscopy tools for analyzing single biomolecular complexes
Review - Dudko, O., Mathé, J. and Meller A. (2010) Nanopore Force Spectroscopy tools for analyzing single biomolecular complexes, In Methods in Enzymology Ed. Walter, N.
Analyzing Individual Biomolecules Using Nanopores
Review - Wanunu, M. Soni, G. and A. Meller. Analyzing Individual Biomolecules Using Nanopores, in the “Handbook of Nanophysics” K.D. Sattler (Ed), 2010 Taylor & Francis Group.
2000-2009
2007-2009
Valencia-Burton, M. Shah, A. Sutin, J. Borogovac, A., McCullough, R. Cantor, C.R. Meller, A. and Broude, N.E. (2009) Spatiotemporal patterns and transcription kinetics of induced RNA in single bacterial cells, Proc. Natl. Acad. Sci. U.S.A. 106, 16399-16404.
Wanunu, M. Sutin, J. and A. Meller (2009) DNA Profiling Using Solid-State Nanopores: Detection of DNA-Binding Molecules, Nano Letters, 9, 3498-3502.
Wanunu, M. Sutin, J. McNally, B. Chow, A. and A. Meller (2008) DNA Translocation Governed by Interactions with Solid State Nanopores, Biophys. J. 95, 4716-25.
McNally, B. Wanunu, M. and A. Meller (2008) Electro-mechanical unzipping of individual DNA molecules using synthetic sub-2 nm pores, Nano Letters, 8, 3418-22.
Wanunu, M. Chakrabarti, B. Mathé, J. Nelson, D. R. and A. Meller (2008) Orientation-dependent interactions of DNA with an a-Hemolysin Channel, Phys Rev. E, 77, 031904.
Soni G. and A. Meller (2007) Progress Towards ultrafast DNA sequencing using solid state nanopores. Clin. Chem. 3, 1996-01
Kim, M-J., B. McNally, K. Murata and A. Meller (2007) Characteristics of solid-state nanometer pores fabricated using transmission electron microscope (TEM) Nanotechnology, 18, 205302.
Dudko, O., J. Mathé, A. Szabo, A. Meller, and G. Hummer (2007) Extracting kinetics from single-molecule force spectroscopy: Nanopore unzipping of DNA hairpins, Biophys. J., 92, 4188-4195.
Wanunu, M. and A. Meller (2007) Chemically Modified Solid-State
Nanopores. Nano Letters, 7, 1580-85, (also read Research Highlight, Nature, 447, 14 (2007)Hornblower, B., A. Coombs, R.D. Whitaker, A. Kolomeisky, Picone, S.J., A. Meller and M. Akeson (2007) Single-Molecule Analysis of DNA-Protein Complexes Using Nanopores. Nature Methods, 4, 315-317 DOI: 10.1038/nmeth1021.
Edel, J., J. Eid and A. Meller (2007) Accurate single molecule FRET efficiency determination for surface immobilized DNA using maximum likelihood calculated lifetimes. J. Phys. Chem B., 111, DOI: 10.1021/jp066530k.
2000-2006
Kim, M-J., M. Wanunu, C.D. Bell and A. Meller (2006) Rapid Fabrication of Uniform Size Nanopores and Nanopore Arrays for Parallel DNA Analysis. Adv. Mater. 18, 3149-3153.
Jan Bonthuis, D., J. Zhang, B. Hornblower, J. Mathé, B.I. Shklovskii and A. Meller (2006) Self-Energy-Limited Ion Transport in Subnanometer Channels. Phys. Rev. Lett. 97, 128104.
Mathé, J., A. Arinstein, Rabin Y. and A. Meller (2006) Equilibrium and irreversible unzipping of DNA in a nanopore, Europhys. Lett. 73 (1), 128-134.
Viasnoff, V., A. Meller and H. Isambert (2006) DNA nanomechanical switches under folding kinetics control. Nano Letters, 6, 101-104.
Sabanayagam, C.R., J.S. Eid and A. Meller (2005) Long time scale blinking kinetics of cyanine fluorophores conjugated to DNA and its effect on Förster resonance energy transfer. J. Chem. Phys. 123, 224708.
Mathé, J., A. Aksimentiev, D.R. Nelson, K. Schulten and A. Meller (2005) Orientation discrimination of single-stranded DNA inside the alpha-hemolysin membrane channel. Proc. Natl. Acad. Sci. U.S.A. 102, 12377-12382.
Sabanayagam, C.R., J.S. Eid and A. Meller (2005) Using fluorescence resonance energy transfer to measure distances along individual DNA molecules: Corrections due to nonideal transfer. J. Chem. Phys. 122, 061103.
Mathé, J., H. Visram, V. Viasnoff, Y. Rabin and A. Meller (2004) Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 87, 3205-3212.
Sabanayagam, C.R., J.S. Eid and A. Meller (2004) High-throughput scanning confocal microscope for single molecule analysis. Appl. Phys. Lett. 84, 1216-1218.
Bates, M., M. Burns and A. Meller (2003) Dynamics of single DNA molecules actively controlled inside a membrane channel. Biophys. J. 84, 2366-2372.
Meller, A., L. Nivon and D. Branton (2001) Voltage-Driven DNA Translocations through a Nanopore. Phys. Rev. Lett. 86, 3435-3438.
Meller, A. and D. Branton (2002) Single molecule measurements of DNA transport through a nanopore. Electrophoresis 23, 2583-2591.
Meller, A., L. Nivon, E. Brandin, J. Golovchenko and D. Branton (2000) Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. U.S.A. 97, 1079-1084.
Reviews
Review - Dudko, O. and Meller, A. (2009) Probing Biomolecular Interactions Using Nanopore Force Spectroscopy. In Encyclopedia of Analytical Chemistry, Meyers, R. A., Ed. John Wiley & Sons Ltd.
Review - Wanunu, M. G. Soni and A. Meller (2009) Single molecule studies of nucleic acids interactions using nanopores, In: Handbook of Single-Molecule Biophysics, Springer, in press
Review - Branton D. et. al. (2008) The potential and challenges of nanopore sequencing, Nature Biotechnology 26, 1146-53.
Review - Wanunu, M. and A. Meller (2008) Single Molecule Analysis of Nucleic Acids and DNA-protein Interactions using Nanopores, Laboratory Manual on Single Molecules, Eds. P. Selvin and T. Ha, Cold Spring Harbor Press.
Review - Lee, J.W. and A. Meller (2006). Rapid DNA Sequencing by Direct Nanoscale Reading of Nucleotide Bases on Individual DNA chains. In: Perspectives in Bioanalysis. K. Mitchelson (Ed.), Elsevier.
Review - Meller, A. (2003) Dynamics of polynucleotide transport through nanometre-scale pores. J. Phys.: Condens. Matter 15, R581-R607.
Review - Branton, D. and A. Meller (2002) Using nanopores to discriminate between single molecules of DNA. 177-185 In: Structure and Dynamics of Confined Polymers, Kluwer Academic Publishers.
1992-1999
1992-1999
Meller, A., J. Stavans, T. Gisler, and D.A. Weitz. (1999) Viscoelasticity of Depletion-Induced Gels in Emulsion-Polymer Systems. Langmuir 15:1918-1922.
Tlusty, T., A. Meller, and R. Bar-Ziv. (1998) Optical gradient forces of strongly localized fields. Phys. Rev. Lett. 81:1738-1741.
Meller, A., R. Bar-Ziv, T. Tlusty, E. Moses, J. Stavans and S.A. Safran. (1998). Localized dynamic light scattering: A new approach for dynamic measurements in optical microscopy. Biophys. J. 74:1541-1548.
Bar-Ziv, R., A. Meller, T. Tlusty, E. Moses, J. Stavans and S.A. Safran (1997). Localized Dynamic Light Scattering: Probing Single Particle Dynamics at the Nanoscale. Phys. Rev. Lett. 78:154-157.
Meller, A. and J. Stavans (1996). Stability of Emulsions with Nonadsorbing Polymers. Langmuir 12:301-304
Steiner, U., A. Meller, and J. Stavans (1995). Entropy Driven Phase Separation in Binary Emulsions. Phys. Rev. Lett. 74:4750-4753.
Meller, A. and J. Stavans. (1992). Glass transition and phase diagrams of strongly interacting binary colloidal mixtures. Phys. Rev. Lett. 68:3646-3649.